

FISH (Fluorescence In Situ Hybridization)
Fluorescence In Situ Hybridization (FISH) is a molecular biology technique used to visualize and locate specific nucleic acid sequences within cells or tissue samples.
Fluorescence In Situ Hybridization (FISH) is a molecular biology technique used to visualize and locate specific nucleic acid sequences within cells or tissue samples. It is a powerful tool in genetics and cytogenetics research and is commonly employed in clinical diagnostics and cancer research.
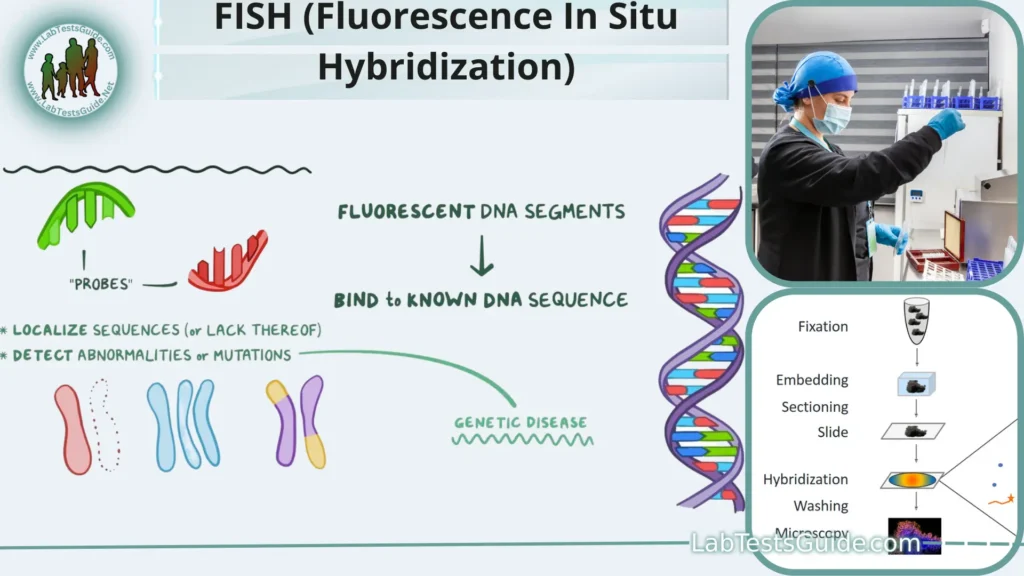
Key Points:
- Molecular Biology Technique – FISH is a molecular biology technique used to study and visualize specific DNA or RNA sequences within cells or tissues.
- DNA/RNA Probes – It employs DNA or RNA probes labeled with fluorescent markers to bind and identify target sequences.
- Complementary Base Pairing – FISH relies on the complementary base pairing between the probe and the target sequence for detection.
- Visualization – Fluorescent signals emitted by the labeled probes are visualized under a fluorescence microscope.
- Genetic Research – FISH is widely used in genetic research to map genes and study genome organization.
- Chromosomal Abnormalities – It helps detect chromosomal abnormalities like deletions, amplifications, and translocations.
- Cancer Research – FISH is essential in cancer research for identifying genetic markers associated with cancer.
- Sample Types – FISH can be applied to various sample types, including cells, tissue sections, and organisms.
- Chromosome Number – It can determine the chromosome number and ploidy levels within cells.
- Prenatal Diagnostics – FISH is used in prenatal diagnostics to detect chromosomal abnormalities in developing fetuses.
- Preimplantation Genetic Diagnosis (PGD) – It plays a role in PGD during in vitro fertilization to select healthy embryos.
- Multicolor FISH (M-FISH) – Multiple probes with different fluorescent labels can be used simultaneously for more complex analysis.
- Spectral Karyotyping (SKY) – SKY is an advanced FISH technique that visualizes all human chromosomes in a single experiment.
- FISH-ES – FISH can identify and quantify microbial species in environmental samples for microbial ecology studies.
- Microbial Communities – It helps study microbial communities in various ecosystems.
- Genome Organization – FISH has revolutionized our understanding of genome organization and structure.
- Probe Design – Successful FISH requires careful probe design to target specific sequences of interest.
- Hybridization Optimization – Optimization of hybridization conditions is crucial for accurate results.
- Specialized Equipment – FISH relies on specialized equipment like fluorescence microscopes for analysis.
- Versatile Tool – FISH is a versatile and powerful tool with applications in genetics, cytogenetics, and clinical diagnostics.
Definition and Overview:
Fluorescence In Situ Hybridization (FISH) is a molecular biology technique that uses fluorescently labeled DNA or RNA probes to locate and visualize specific genetic sequences within cells or tissues. It has wide-ranging applications in genetics, cytogenetics, and clinical diagnostics. FISH revolutionizes our ability to study DNA and RNA at the cellular level by providing precise localization and identification of target sequences.
Historical Background:
Fluorescence In Situ Hybridization (FISH) has its origins in the mid-20th century. The technique evolved from earlier methods used to study chromosomal structure and organization. Here’s a brief historical overview:
- 1960s-1970s : Early studies involved using radioactively labeled DNA probes to detect complementary sequences in chromosomes. However, radioactivity posed safety concerns and limited the practicality of the method.
- 1970s-1980s : Researchers began exploring non-radioactive alternatives, leading to the development of FISH. Pioneering work by scientists like Joe Gall and Mary-Lou Pardue in the 1970s laid the foundation for the technique.
- 1980s : The term “Fluorescence In Situ Hybridization” was coined, and FISH gained popularity as a powerful tool for studying genetic material. It provided a safer and more versatile alternative to radioactive probes.
- 1986 : The first publication on FISH using fluorescent probes was authored by J.W. Pinkel and D. Gray, marking a significant milestone in the technique’s development.
- 1990s-Present : FISH continued to advance rapidly, with the development of multicolor FISH (M-FISH), spectral karyotyping (SKY), and various specialized applications in genetics, cytogenetics, and clinical diagnostics.
Today, FISH remains a cornerstone of genetic research and diagnostics, enabling researchers and clinicians to explore the intricacies of DNA and RNA localization and organization within cells and tissues. Its historical evolution reflects the ongoing innovation in molecular biology and genetic analysis techniques.
Purpose of FISH:
- Localization and Mapping: FISH allows researchers to precisely locate and map the positions of genes, genetic markers, or specific DNA/RNA sequences within chromosomes. This is crucial for understanding the organization of genetic material.
- Cytogenetic Analysis: In cytogenetics, FISH is used to detect and analyze chromosomal abnormalities such as translocations, deletions, duplications, and inversions. It plays a key role in diagnosing genetic disorders.
- Cancer Research and Diagnosis: FISH is widely used in cancer research and diagnosis. It helps identify genetic alterations, such as gene amplifications or chromosomal rearrangements, associated with various cancers. This information guides treatment decisions and prognosis assessment.
- Prenatal and Preimplantation Diagnostics: FISH is employed in prenatal testing to detect chromosomal abnormalities in developing fetuses. In preimplantation genetic diagnosis (PGD), it assesses embryos for genetic abnormalities before implantation during in vitro fertilization (IVF).
- Microbiology and Environmental Studies: FISH-ES (Environmental Sample) is used to identify and quantify specific microbial species in environmental samples. It contributes to the study of microbial diversity and ecosystems.
- Gene Expression Analysis: FISH helps researchers investigate gene expression patterns by visualizing RNA molecules within cells. This is valuable for understanding how genes are regulated and how they contribute to cellular functions.
- Neurobiology and Developmental Biology: FISH is applied in neurobiology to study gene expression patterns in the brain and neuronal tissues. In developmental biology, it provides insights into gene expression during embryogenesis and tissue differentiation.
- Plant Genetics: FISH is used in plant genetics to study chromosomal organization, genetic diversity, and the identification of specific genes or markers.
- Clinical Diagnostics: FISH is routinely used in clinical laboratories for diagnosing genetic disorders, detecting cancer-related genetic alterations, and guiding treatment decisions.
- Research Tool: FISH is a versatile research tool in various biological fields, allowing scientists to investigate the genetic and molecular basis of diverse biological processes and diseases.
Principle of FISH:
The principle of FISH revolves around the specific binding of fluorescently labeled DNA or RNA probes to complementary target sequences within the genetic material of cells or tissues. Here’s how it works:
- Probe Design : Specific DNA or RNA probes are designed to be complementary to the sequences of interest. These probes are usually relatively short, typically 20-30 nucleotides in length, and are chosen to match the target DNA or RNA.
- Fluorescent Labeling : The DNA or RNA probes are labeled with fluorescent molecules, also known as fluorophores. These fluorophores emit light when exposed to specific wavelengths of light.
- Sample Preparation : The target cells or tissue sections are prepared for FISH. This may involve fixation to immobilize cellular structures and denaturation to expose the target DNA or RNA.
- Hybridization : The labeled probes are applied to the prepared sample. The probes will seek out and bind (hybridize) to their complementary sequences within the sample’s DNA or RNA.
- Washing : After hybridization, the sample is washed to remove any unbound or nonspecifically bound probes. This step reduces background fluorescence.
- Detection : The sample is examined under a fluorescence microscope equipped with appropriate filters. When exposed to specific wavelengths of light, the bound probes emit fluorescent signals at their characteristic wavelengths.
- Imaging and Analysis : A camera attached to the microscope captures images of the fluorescent signals. These images are then analyzed to determine the location, quantity, and distribution of the target sequences within the sample.
FISH Applications:
Fluorescence In Situ Hybridization (FISH) has a wide range of applications in genetics, cytogenetics, and clinical diagnostics due to its ability to visualize and locate specific DNA or RNA sequences within cells or tissues. Here are some key applications:
- Gene Mapping : FISH is used to map the location of specific genes on chromosomes, helping researchers understand the genetic basis of various traits and diseases.
- Chromosomal Abnormality Detection : It plays a crucial role in identifying chromosomal abnormalities, including deletions, duplications, translocations, and inversions, which are associated with genetic disorders.
- Cancer Research : FISH is extensively used in cancer research to detect genetic alterations such as gene amplifications, deletions, and rearrangements, aiding in the understanding of cancer development and progression.
- Prenatal Diagnostics : In prenatal testing, FISH can identify chromosomal abnormalities in developing fetuses, providing valuable information for expectant parents and healthcare providers.
- Preimplantation Genetic Diagnosis (PGD) : FISH is employed during in vitro fertilization (IVF) to screen embryos for chromosomal abnormalities before implantation, reducing the risk of genetic disorders in offspring.
- Microbial Ecology : FISH-ES (Fluorescent In Situ Hybridization for Environmental Samples) is used to identify and quantify specific microbial species in environmental samples, aiding in the study of microbial communities.
- Species Identification : FISH can be used to differentiate and identify species or strains of microorganisms, making it valuable in microbiology and infectious disease research.
- Telomere Analysis : It helps researchers study telomere length and stability, which is associated with aging and cancer.
- Molecular Cytogenetics : FISH complements conventional cytogenetic techniques, providing higher resolution and precision in the analysis of chromosomes and genetic material.
- Multicolor FISH (M-FISH) : M-FISH enables the simultaneous visualization of multiple DNA targets, allowing for complex chromosomal analyses and the identification of complex rearrangements.
- Spectral Karyotyping (SKY) : SKY is an advanced FISH technique that can visualize all human chromosomes in a single hybridization experiment, aiding in the diagnosis of chromosomal abnormalities.
- FISH Combined with Immunohistochemistry (FISH-IHC) : This technique combines FISH with immunohistochemistry to simultaneously detect genetic and protein markers, enhancing the understanding of disease mechanisms.
- Bacterial and Viral Detection : FISH is used to detect and identify specific bacterial or viral pathogens in clinical samples, contributing to infectious disease diagnosis.
- Assisted Reproduction : FISH helps assess the genetic health of embryos in assisted reproductive technologies, ensuring the selection of embryos with normal chromosomal content.
- Plant Genetics : FISH is applied in plant genetics to study chromosomal organization, genetic diversity, and the identification of specific genes or markers.
- Forensic Analysis : FISH can be employed in forensic science to identify specific DNA sequences, aiding in criminal investigations and paternity testing.
- Comparative Genomic Hybridization (CGH) : FISH is used in CGH to compare the DNA copy number variations between normal and diseased cells, facilitating the identification of genetic factors in diseases.
Required Sample and its Preparation:
The choice of sample and its proper preparation are critical steps in conducting FISH experiments. Different types of samples require specific preparation procedures. Here are the key considerations for sample selection and preparation:
1. Sample Types:
- Cell Cultures : For cultured cells, samples are typically grown on coverslips or culture dishes. This method allows for controlled conditions and easy preparation.
- Tissue Sections : In clinical and histological studies, tissue sections on slides are often used. Tissues can be fresh or formalin-fixed and paraffin-embedded (FFPE).
2. Sample Selection:
- Ensure that the sample contains the target DNA or RNA sequences of interest. This might involve selecting specific cell types, regions, or tissues for analysis.
3. Sample Fixation:
- Fixation is essential to preserve cellular structures and prevent sample degradation.
- For cultured cells, fixation is usually done with formaldehyde or paraformaldehyde.
- For tissue samples, formalin fixation and paraffin embedding (FFPE) are common in clinical settings. Alternatively, fresh-frozen tissue samples can be used.
4. Slide Preparation:
- Cells or tissue sections are often mounted onto glass slides for FISH.
- In the case of FFPE samples, the tissue is thinly sliced into sections, and the sections are placed on slides.
5. Deparaffinization (if using FFPE):
- If using FFPE samples, the paraffin wax must be removed. This is done through a series of xylene and alcohol washes.
6. Denaturation:
- Denaturation is a crucial step to make the DNA or RNA accessible for probe binding. This is typically done by exposing the sample to high temperatures (e.g., 80-90°C) in a denaturation buffer.
7. Permeabilization (optional):
- In some cases, permeabilization of the cell membrane may be necessary to allow probe penetration, especially for tissue samples. This step is usually done with a detergent solution.
8. Probe Hybridization:
- After denaturation and, if needed, permeabilization, the fluorescently labeled probes are applied to the sample. These probes are typically mixed with a hybridization buffer and allowed to incubate with the sample.
9. Post-Hybridization Washes:
- Unbound probes are removed through a series of stringent washes. This step helps reduce background fluorescence.
10. Counterstaining and Mounting: – To visualize cellular structures and nuclei, counterstaining with dyes like DAPI (4′,6-diamidino-2-phenylindole) may be performed. – Finally, the sample is mounted with an anti-fade mounting medium to preserve fluorescence for imaging.
Specialized Equipment:
Fluorescence In Situ Hybridization (FISH) is a technique that requires specialized equipment to visualize and analyze the fluorescent signals emitted by labeled probes binding to target DNA or RNA sequences. Here are some of the key pieces of specialized equipment used in FISH:
- A fluorescence microscope is the central piece of equipment for FISH.
- It has the capability to excite fluorophores with specific wavelengths of light and capture emitted fluorescence signals.
- The microscope is equipped with filters that selectively allow the passage of fluorescence emissions while blocking other wavelengths.
- High-quality optics and objectives are essential for clear and detailed imaging.
- Fluorescence microscopes are equipped with sets of excitation and emission filters specific to the fluorophores used in FISH.
- Excitation filters allow only the excitation wavelength to reach the sample.
- Emission filters allow the emitted fluorescence to pass through for detection.
- A digital camera attached to the microscope captures high-resolution images of the sample.
- Specialized imaging software controls camera settings, image acquisition, and data storage.
- The camera’s sensitivity and resolution are crucial for obtaining quality images.
- FISH requires precise and controlled illumination.
- Many fluorescence microscopes use arc lamps, lasers, or LEDs as light sources to excite fluorophores.
- Light intensity can be adjusted to optimize image quality and reduce photobleaching.
- Specialized software is used for image processing, analysis, and quantification of FISH results.
- It aids in tasks such as signal counting, localization, and measurement of fluorescence intensity.
- Advanced software may also facilitate 3D image reconstruction and co-localization analysis.
- Maintaining a stable temperature is crucial during FISH experiments, especially during the hybridization step.
- Specialized heating stages or incubation chambers can be integrated into the microscope.
- Microscopes are equipped with a range of objective lenses with varying magnifications and numerical apertures.
- The choice of objective lens depends on the specific FISH experiment and sample type.
- For samples with three-dimensional structures, accessories like motorized stages and focus controllers allow the capture of Z-stack images at different focal planes.
- These images can be reconstructed to create 3D representations of the sample.
- These components hold and position the glass slides containing the FISH samples for imaging.
- Motorized stages enable precise movement and automation of image capture.
- Specialized reagents may be used to reduce background fluorescence and preserve the fluorophores’ signals during imaging.
- In some cases, a darkroom or low-light environment may be used to minimize external light interference during imaging.
Procedure of FISH:
Fluorescence In Situ Hybridization (FISH) is a multi-step molecular biology technique used to visualize and locate specific DNA or RNA sequences within cells or tissues. The procedure involves several key steps, from sample preparation to image analysis. Here’s a step-by-step overview of the FISH procedure:
1. Sample Selection and Preparation:
- Choose the appropriate sample type (e.g., cultured cells, tissue sections).
- Fix the sample to preserve cellular structures and prevent degradation.
- For clinical samples, prepare formalin-fixed and paraffin-embedded (FFPE) sections or fresh-frozen samples.
- Mount cells or tissue sections on glass slides.
2. Deparaffinization (if using FFPE samples):
- If using FFPE samples, remove paraffin wax through a series of xylene and alcohol washes.
3. Denaturation:
- Expose the sample to high temperatures (usually 80-90°C) in a denaturation buffer to make the DNA or RNA accessible for probe binding.
- This step can involve heating the sample on a hotplate or in an oven, followed by a brief cooling period.
4. Probe Hybridization:
- Prepare the fluorescently labeled DNA or RNA probes specific to the target sequences.
- Mix the probes with a hybridization buffer to create a probe solution.
- Apply the probe solution to the sample on the slide, ensuring even distribution.
- Incubate the sample at a specific temperature (typically 37-42°C) to allow the probes to bind to their complementary sequences within the sample.
5. Post-Hybridization Washes:
- Remove unbound probes through a series of stringent washes with buffers.
- These washes help reduce background fluorescence and increase signal specificity.
6. Counterstaining (optional):
- To visualize cellular structures or nuclei, apply a counterstain, often using a dye like DAPI (4′,6-diamidino-2-phenylindole).
- Counterstaining can provide context for the localization of target sequences within cells.
7. Mounting:
- Apply an anti-fade mounting medium to preserve fluorescence and protect the sample from photobleaching.
8. Microscopy and Imaging:
- Examine the sample under a fluorescence microscope equipped with appropriate filters for the fluorophores used in the probes.
- Excite the fluorescent labels with specific wavelengths of light and capture images of the emitted fluorescence.
9. Image Analysis:
- Analyze the acquired images to determine the location, quantity, and distribution of the target sequences within the sample.
- Specialized software may be used for image processing and quantification.
10. Data Interpretation: – Interpret the results based on the distribution and intensity of fluorescence signals. – Correlate the findings with the research or diagnostic objectives.
11. Documentation and Reporting: – Document the experimental details and results for publication or clinical reporting.
Probe Design:
Probe design is a critical step in the Fluorescence In Situ Hybridization (FISH) technique. The design ensures that the probes are specific to the target DNA or RNA sequences of interest, enabling accurate visualization and localization. Here’s an overview of the key considerations and steps in probe design for FISH:
1. Target Sequence Selection:
- Identify the specific DNA or RNA sequence you want to detect or visualize within the sample. This could be a gene, a chromosomal region, or a non-coding RNA.
2. Probe Length:
- Probes are typically short, usually 20-30 nucleotides in length. Longer probes may be used for larger targets.
- Ensure that the probe length is appropriate for the target sequence to enhance binding specificity.
3. Sequence Specificity:
- Design the probe to be highly complementary to the target sequence. Avoid sequences with repetitive elements or regions prone to cross-hybridization.
4. GC Content and Melting Temperature (Tm):
- Calculate the GC content and Tm of the probe to ensure stable hybridization. Probes with balanced GC content and appropriate Tm values are more likely to bind specifically to the target.
5. Avoid Secondary Structures:
- Check for potential self-complementarity or secondary structures within the probe sequence that may hinder hybridization efficiency.
6. Labeling:
- Choose a method for labeling the probe with fluorescent molecules (fluorophores). Common labels include fluorescein, Cy3, Cy5, and Alexa Fluor dyes.
- Label the probe at the 5′ or 3′ end or incorporate multiple labels along its length for increased signal intensity.
7. Quenching:
- Consider using quenchers to reduce background fluorescence. Quenchers absorb emitted light from fluorophores and prevent it from interfering with signal detection.
8. Avoid Polymorphisms:
- Check for single-nucleotide polymorphisms (SNPs) or mutations in the target sequence that might affect probe binding. Adjust the probe design accordingly.
9. Test Probes In Silico:
- Utilize bioinformatics tools and software to perform in silico analysis of probe sequences, including specificity, melting temperature, and secondary structures.
10. Validation: – Experimentally validate the probe’s specificity and efficiency through hybridization assays using control samples with known target sequences.
11. Positive and Negative Controls: – Include positive controls with known target sequences and negative controls lacking the target sequence in your FISH experiments.
12. Multiple Probes (Optional): – For complex analyses or multicolor FISH (M-FISH), design and label multiple probes to simultaneously target different sequences within the sample.
13. Optimization: – Fine-tune hybridization conditions, such as temperature and probe concentration, to achieve optimal results.
Effective probe design is essential for the success of FISH experiments, ensuring specific and reliable detection of target sequences within cells or tissues. Careful consideration of sequence specificity, length, labeling, and in silico analysis contributes to accurate and meaningful FISH results.
Hybridization Process:
The hybridization process in Fluorescence In Situ Hybridization (FISH) is a critical step where fluorescently labeled DNA or RNA probes bind to their complementary target sequences within the sample. This process allows researchers to visualize and locate specific genetic sequences. Here’s a detailed overview of the hybridization process in FISH:
1. Probe Preparation:
- Start with the preparation of the DNA or RNA probes specific to the target sequences of interest.
- Label the probes with fluorescent molecules (fluorophores) for subsequent detection.
2. Sample Preparation:
- Prepare the sample containing the cells or tissue sections to be analyzed using FISH.
- If using formalin-fixed and paraffin-embedded (FFPE) samples, deparaffinize and rehydrate them.
- Denature the sample to expose the target DNA or RNA strands.
3. Probe Application:
- Apply the fluorescently labeled probes to the denatured sample.
- Mix the probe solution with a hybridization buffer to create the hybridization mixture.
- Ensure the probe solution covers the entire sample evenly.
4. Hybridization Incubation:
- Temperature: Typically 37-42°C, depending on the probe and target sequences.
- Duration: The incubation time can vary but usually ranges from several hours to overnight.
5. Probe Binding:
- During incubation, the probes seek out and bind (hybridize) to their complementary sequences within the sample’s DNA or RNA.
- The binding occurs through hydrogen bonding between complementary base pairs (A-T and G-C).
6. Stringency Control:
- Stringency control is essential to ensure specific probe-target binding.
- Stringency is controlled by adjusting temperature, salt concentration, and wash conditions.
- Higher stringency conditions reduce non-specific binding.
7. Post-Hybridization Washes:
- After hybridization, wash the sample to remove unbound probes and minimize background fluorescence.
- Use buffers with specific stringency levels to achieve the desired specificity.
8. Counterstaining (Optional):
- To visualize cellular structures, nuclei, or other components, counterstaining can be performed using dyes like DAPI (4′,6-diamidino-2-phenylindole).
9. Mounting:
- Apply an anti-fade mounting medium to preserve the fluorescence and protect the sample from photobleaching.
10. Microscopy and Imaging: – Examine the sample under a fluorescence microscope equipped with appropriate filters for the fluorophores used in the probes. – Illuminate the sample with specific wavelengths of light to excite the fluorescent labels. – Capture images of the emitted fluorescent signals.
11. Image Analysis: – Analyze the acquired images to determine the location, quantity, and distribution of the target sequences within the sample. – Use specialized software for image processing and quantification if necessary.
Detection and Imaging:
After the hybridization process, the next key steps in Fluorescence In Situ Hybridization (FISH) involve detecting and imaging the fluorescent signals produced by the labeled probes binding to their target sequences within the sample. Here’s an overview of the detection and imaging process in FISH:
1. Fluorescence Microscopy:
- Use a fluorescence microscope equipped with appropriate filters for the specific fluorophores used to label the FISH probes.
2. Illumination:
- Illuminate the sample with specific wavelengths of light that excite the fluorescent labels (fluorophores).
- Each fluorophore has a unique excitation wavelength.
3. Emission of Fluorescent Signals:
- When excited by the appropriate wavelength of light, the fluorophores emit fluorescent signals at their characteristic wavelengths.
4. Filter Selection:
- Use fluorescence filters to selectively capture emitted light from the fluorophores while blocking other wavelengths.
- Filter sets consist of excitation and emission filters that match the fluorophore’s spectral characteristics.
5. Image Acquisition:
- Capture digital images of the sample using a camera attached to the fluorescence microscope.
- Take multiple images at different focal planes if the sample is three-dimensional.
6. Exposure Time and Sensitivity:
- Adjust exposure times and sensitivity settings to optimize image quality and signal-to-noise ratio.
7. Multichannel Imaging (Optional):
- If using multiple fluorophores to label different probes, capture images for each channel (fluorophore) separately.
- This allows you to differentiate and visualize multiple target sequences simultaneously.
8. Image Stacking (Z-Stack):
- If imaging a three-dimensional sample, capture a series of images at different focal planes.
- Combine these images to create a stacked or 3D image, providing a comprehensive view of the sample.
9. Image Processing and Analysis:
- Process the acquired images using specialized software for tasks such as deconvolution, background subtraction, and noise reduction.
- Analyze the images to quantify signal intensity, localization, and distribution of the target sequences within the sample.
10. Visualization and Interpretation: – Visualize the fluorescent signals in the images, which represent the locations of the target DNA or RNA sequences. – Interpret the results based on the distribution and intensity of the signals, correlating them with the research or diagnostic objectives.
11. Data Storage and Documentation: – Store and archive the acquired images and associated data for future reference and reporting. – Document the experimental details, including imaging settings and conditions.
Advantages and Limitations:
Advantages:
- High Specificity : FISH offers exceptional specificity, allowing for the precise detection and localization of target DNA or RNA sequences within cells or tissues.
- Visualization of Genetic Material : It provides visual, direct, and in situ visualization of genetic material, allowing researchers to see where specific genes or sequences are located within the cell.
- Cytogenetic Analysis : FISH is invaluable in cytogenetics for studying chromosomal abnormalities, including deletions, duplications, translocations, and inversions, aiding in the diagnosis of genetic disorders.
- Cancer Research : FISH plays a crucial role in cancer research by identifying genetic alterations associated with tumor development and progression.
- Prenatal and Preimplantation Diagnostics : FISH is used in prenatal and preimplantation genetic diagnostics to detect chromosomal abnormalities in fetuses and embryos, reducing the risk of genetic disorders.
- Microbial Ecology : FISH-ES allows the identification and quantification of microbial species in environmental samples, enhancing our understanding of microbial communities.
- Multicolor FISH (M-FISH) : M-FISH enables the simultaneous visualization of multiple target sequences, facilitating complex chromosomal analyses.
- Spectral Karyotyping (SKY) : SKY provides a comprehensive view of all human chromosomes in a single experiment, aiding in the diagnosis of chromosomal abnormalities.
- Research Tool : FISH is a versatile research tool used in various fields, including genetics, microbiology, developmental biology, and neuroscience.
- Assisted Reproduction : It assists in selecting embryos with normal chromosomal content during in vitro fertilization (IVF) procedures.
Limitations:
- Limited Resolution : FISH has limited resolution, making it challenging to detect small-scale genetic changes or subtle rearrangements.
- Background Signal : Non-specific binding of probes or autofluorescence can lead to background signals, potentially affecting result accuracy.
- Probe Design : Designing specific and effective probes can be complex, requiring careful consideration of factors such as sequence specificity and melting temperature.
- Hybridization Optimization : Achieving optimal hybridization conditions can be time-consuming and may require experimentation to reduce non-specific binding.
- Sample Complexity : Complex samples with multiple genetic elements or structural variations can complicate FISH analysis.
- Limited Quantification : FISH is primarily a qualitative technique, and quantification may be challenging, especially for low-level target sequences.
- Cell Permeability : In some samples, achieving probe penetration may be difficult due to impermeable cell membranes or tissue structures.
- Cost and Resources : FISH can be resource-intensive, requiring specialized equipment and fluorescently labeled probes, which can be expensive.
- Sample Preparation Variability : Variability in sample preparation can impact FISH results, requiring strict adherence to protocols.
- Expertise Required : FISH analysis requires trained personnel with expertise in probe design, experimental setup, and image analysis.
Variants and Advanced Techniques:
Fluorescence In Situ Hybridization (FISH) has evolved over the years, leading to various variants and advanced techniques that offer enhanced capabilities and applications. Here are some notable variants and advanced techniques of FISH:
- M-FISH uses a combination of differently labeled probes to simultaneously visualize multiple target sequences within a single sample.
- Each chromosome or target is assigned a unique color, enabling the identification of complex chromosomal abnormalities.
- SKY is an advanced form of FISH that employs a spectrally distinct set of fluorophores to visualize all human chromosomes in a single hybridization experiment.
- It allows for the detection of complex chromosomal rearrangements and structural abnormalities.
- FISH-FACS combines FISH with flow cytometry to sort and analyze cells based on their DNA content and specific DNA sequences.
- It is valuable in studying cell populations with heterogeneous genomes, such as cancer cells.
- FISH-IHC combines FISH with immunohistochemistry to simultaneously detect genetic and protein markers within the same tissue section.
- This technique provides insights into the correlation between genetic alterations and protein expression in disease states.
- CGH, including array CGH, is a high-resolution technique that compares the DNA copy number variations between normal and diseased cells.
- It is used to identify genomic imbalances associated with diseases, including cancer.
- Q-FISH involves the use of specialized probes and image analysis techniques to quantitatively measure the copy number and telomere length of specific DNA sequences.
- It is applied in aging research, cancer studies, and telomere-related diseases.
- LNA probes incorporate modified nucleotides called locked nucleic acids, which enhance probe binding specificity and stability.
- LNAs are particularly useful in challenging FISH applications requiring high stringency conditions.
- RNA FISH is used to visualize and localize specific RNA molecules within cells.
- It provides insights into gene expression patterns, RNA localization, and the study of non-coding RNAs.
- Digital FISH involves the use of nanoscale materials and advanced imaging techniques to achieve higher sensitivity and resolution in FISH experiments.
- It is used in single-molecule FISH (smFISH) for single-cell RNA analysis.
- Fiber FISH is used to visualize and measure the length and distribution of extended DNA or chromatin fibers.
- It aids in the study of chromosomal organization and structural changes.
Research and Clinical Applications:
Fluorescence In Situ Hybridization (FISH) has a wide range of research and clinical applications, spanning various fields of biology and medicine. Here are some key research and clinical applications of FISH:
Research Applications:
- Gene Mapping and Localization: FISH is used to map the location of specific genes on chromosomes, helping researchers understand the genetic basis of traits and diseases.
- Cytogenetics: FISH is a cornerstone technique in cytogenetics, allowing the detection of chromosomal abnormalities such as translocations, deletions, duplications, and inversions.
- Cancer Research: FISH is extensively employed in cancer research to identify genetic alterations associated with various cancers. It aids in understanding cancer development, prognosis, and treatment options.
- Genomic Imbalances: Comparative Genomic Hybridization (CGH) and array CGH use FISH to compare DNA copy number variations between normal and diseased cells, revealing genomic imbalances in diseases like cancer and developmental disorders.
- Telomere Analysis: FISH is used to study telomere length and stability, which have implications for aging, cancer, and genetic diseases.
- Microbial Ecology: FISH-ES is used to identify and quantify specific microbial species in environmental samples, enhancing the study of microbial communities and their ecological roles.
- Molecular Cytogenetics: FISH complements traditional cytogenetic techniques by providing higher resolution in the analysis of chromosomes and genetic material.
- Plant Genetics: FISH is applied in plant genetics to study chromosomal organization, genetic diversity, and the identification of specific genes or markers.
- Neuroscience: FISH is used to study gene expression patterns in the brain and neuronal tissues, providing insights into neurodevelopment and neurological disorders.
- Developmental Biology: FISH is applied to study the spatiotemporal expression patterns of genes during development, shedding light on embryogenesis and tissue differentiation.
Clinical Applications:
- Clinical Cytogenetics: FISH is routinely used in clinical laboratories to diagnose genetic disorders, such as Down syndrome (Trisomy 21), Prader-Willi syndrome, and Angelman syndrome.
- Cancer Diagnosis: FISH is employed to detect genetic alterations associated with various cancers, aiding in the diagnosis and prognosis of malignancies.
- Prenatal Testing: FISH is used in prenatal diagnostics to identify chromosomal abnormalities in developing fetuses, providing valuable information for expectant parents and healthcare providers.
- Preimplantation Genetic Diagnosis (PGD): FISH is applied during in vitro fertilization (IVF) to screen embryos for chromosomal abnormalities before implantation, reducing the risk of genetic disorders in offspring.
- Infectious Disease Diagnosis: FISH is used to detect and identify specific bacterial or viral pathogens in clinical samples, contributing to the diagnosis of infectious diseases.
- Assisted Reproduction: FISH helps assess the genetic health of embryos in assisted reproductive technologies, ensuring the selection of embryos with normal chromosomal content.
- Clinical Oncology: FISH is used to guide cancer treatment decisions by identifying genetic markers and targetable alterations in tumor cells.
- Neuropathology: FISH is used to diagnose and classify brain tumors based on specific genetic alterations, aiding in treatment planning.
- Hematology: FISH is employed in the diagnosis and monitoring of hematological disorders, including leukemia and lymphoma.
- Clinical Microbiology: FISH is used for the rapid identification of pathogens in clinical samples, facilitating timely treatment.
Advanced FISH Techniques:
Advanced Fluorescence In Situ Hybridization (FISH) techniques have been developed to address specific research and diagnostic needs, offering improved sensitivity, resolution, and multiplexing capabilities. Here are some advanced FISH techniques:
- Q-FISH incorporates specialized probes and image analysis to quantitatively measure the copy number and telomere length of specific DNA sequences.
- It is used to study telomere biology, aging, and diseases related to telomere length.
- FISH-FACS combines FISH with flow cytometry, allowing the sorting and analysis of cells based on their DNA content and specific DNA sequences.
- It is valuable for studying heterogeneous cell populations and rare cell types.
- It provides insights into the correlation between genetic alterations and protein expression in disease states.
- Digital FISH employs nanoscale materials and advanced imaging techniques to achieve higher sensitivity and resolution in FISH experiments.
- SR-FISH techniques, such as STORM (Stochastic Optical Reconstruction Microscopy) FISH, PALM (Photoactivated Localization Microscopy) FISH, and dSTORM (direct STORM) FISH, break the diffraction limit, achieving nanoscale resolution.
- They are used to visualize molecular details within cellular structures.
- ST-FISH combines FISH with spatial transcriptomics technologies to simultaneously visualize RNA expression patterns and their spatial distribution in tissue sections.
- It aids in understanding tissue organization and cell-to-cell interactions.
- It helps study chromosomal organization and structural changes.
- RNA FISH allows the visualization and localization of specific RNA molecules within cells, providing insights into gene expression patterns and RNA localization.
- smFISH techniques enable the detection and quantification of individual RNA molecules in single cells, offering high sensitivity and single-cell resolution.
- mFISH techniques utilize multiple probes labeled with distinct fluorophores to simultaneously detect and visualize multiple target sequences within a single sample.
- They are valuable for studying complex genetic interactions and gene expression patterns.
- WCP-FISH uses chromosome-specific paint probes that cover entire chromosomes.
- It aids in the visualization and identification of whole chromosomes or chromosome regions.
1. What is Fluorescence In Situ Hybridization (FISH)?
FISH is a molecular biology technique that uses fluorescently labeled DNA or RNA probes to bind and visualize specific genetic sequences within cells or tissues.
2. How does FISH work?
FISH involves hybridizing fluorescent probes with complementary target DNA or RNA sequences within a sample. When the probes bind to their targets, they emit fluorescence, which is detected and visualized under a fluorescence microscope.
3. What are the primary applications of FISH?
FISH is used for gene mapping, cytogenetics, cancer research, prenatal and preimplantation diagnostics, microbiology, and various other research and clinical applications.
4. What is the difference between DNA FISH and RNA FISH?
DNA FISH involves detecting specific DNA sequences, while RNA FISH detects RNA molecules, providing insights into gene expression patterns.
5. What are the advantages of FISH?
FISH offers high specificity, allowing precise localization of genetic sequences. It is valuable in cytogenetics, cancer research, and prenatal diagnostics, among other applications.
6. What are the limitations of FISH?
FISH has limited resolution, requires specialized probes, and can be resource-intensive. Background signal and probe design challenges are also limitations.
7. What is the role of fluorescence microscopy in FISH?
Fluorescence microscopy is essential in FISH for visualizing and capturing the fluorescent signals emitted by the labeled probes binding to target sequences.
8. Are there advanced FISH techniques?
Yes, advanced FISH techniques include M-FISH, SKY, smFISH, super-resolution FISH, and others that offer improved sensitivity, resolution, and multiplexing capabilities.
9. How is FISH used in cancer diagnosis and research?
FISH is used to identify genetic alterations associated with cancer, aiding in the diagnosis, prognosis, and targeted treatment of various malignancies.
10. Is FISH used in prenatal testing?
Yes, FISH is used in prenatal testing to detect chromosomal abnormalities in developing fetuses, providing information for expectant parents and healthcare providers.
11. What is the difference between FISH and PCR (Polymerase Chain Reaction)?
FISH visualizes specific DNA or RNA sequences within cells or tissues, while PCR amplifies and detects DNA or RNA sequences in vitro.
12. Is FISH applicable to other organisms besides humans?
Yes, FISH can be applied to study genetic sequences and chromosomal structures in a wide range of organisms, including plants, animals, and microorganisms.
13. Can FISH be used for single-cell analysis?
Yes, techniques like smFISH enable the detection and quantification of individual RNA molecules in single cells, providing single-cell resolution.
14. What are some challenges in FISH experiments?
Challenges include probe design, optimization of hybridization conditions, and minimizing background fluorescence. Sample preparation and image analysis can also be demanding.
15. Is FISH widely used in clinical diagnostics?
Yes, FISH is routinely used in clinical laboratories for diagnosing genetic disorders, detecting cancer-related genetic alterations, and guiding treatment decisions.
Conclusion:
In conclusion, Fluorescence In Situ Hybridization (FISH) is a powerful molecular biology technique that has revolutionized our ability to visualize and study specific DNA or RNA sequences within cells and tissues. Its versatility spans various fields, including genetics, genomics, cytogenetics, cancer research, and clinical diagnostics. By harnessing the principles of hybridization and fluorescence microscopy, FISH enables researchers and clinicians to:
- Map and locate genes and genetic sequences on chromosomes.
- Diagnose and understand chromosomal abnormalities and genetic disorders.
- Investigate cancer-related genetic alterations, aiding in diagnosis and treatment.
- Facilitate prenatal and preimplantation genetic testing.
- Study microbial communities and environmental microbiology.
- Analyze gene expression patterns and RNA localization.
- Advance our understanding of neurobiology, developmental biology, and many other areas of biology.
FISH continues to evolve with the development of advanced variants and techniques, offering higher resolution, sensitivity, and multiplexing capabilities. As technology advances, FISH remains a valuable tool for addressing complex biological questions and providing critical insights into the molecular and genetic basis of health and disease.
As researchers and clinicians harness the potential of FISH and its advanced applications, it is likely to continue playing a pivotal role in scientific discovery, clinical diagnosis, and the advancement of personalized medicine in the years to come.
Lab Tests Guide Founder
The website covers a wide range of lab tests, including blood tests, urine tests, stool tests, and imaging tests such as X-rays and CT scans. It also provides information about different health conditions and diseases, as well as tips for maintaining good health.
It's important to note that while labtestsguide.com may provide valuable information about lab tests and their interpretation, it's always best to consult with a healthcare professional if you have any concerns or questions about your lab results. We can provide personalized guidance and advice based on your individual health status and medical history.
Similar Posts

Fluorescence In Situ Hybridization (FISH)
Western Blotting (Immunoblotting)

Chemiluminescence immunoassay (CLIA)

Spectrophotometry (SP)
Leave a reply cancel reply.
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.

Nucleus of Notes, News, and Update
FISH- Definition, Principle, Probes, Procedure, Applications
Table of Contents
Introduction:
The cytogenetic method known as fluorescence in situ hybridization (FISH) was developed in the early 1980s. FISH produces coloured signals that may be seen under a fluorescence microscope by utilizing fluorescent DNA probes to target particular chromosomal regions within the nucleus.
It is a macromolecule recognition approach that is regarded as a new development in the field of cytology for identifying specific DNA sequences, diagnosing genetic diseases, mapping genes, and discovering new oncogenes or genetic abnormalities causing various types of cancer within a cell or tissue sample.
FISH is a simple technique that involves hybridizing a DNA probe to its complementary sequence on chromosomal preparations that have been previously placed on slides. Probes are labeled either directly with fluorescent nucleotides or indirectly with reporter molecules that are then recognized by fluorescent antibodies or other affinity molecules. Finally, microscopy analysis is used to visualize probes and targets in situ.
The main advantage of this visually appealing technique as a combined molecular and cytological approach is its unique capacity to provide an intermediate degree of resolution between DNA analysis and chromosomal investigations, while also keeping information at the single-cell level.
FISH works on the basis of a labeled probe specifically binding to complementary DNA or RNA sequences in a sample. The probe is tagged with a fluorescent dye and can be made of RNA or DNA. When the probe is incorporated into the sample, it binds to complementary sequences, either within the same chromosome or between chromosomes.
The probes are either directly labeled by incorporating a fluorophore or indirectly by incorporating a hapten. Following denaturation, the labeled probe and the target DNA are combined, allowing complementary DNA sequences to anneal. If the probe had been indirectly labeled, it would be necessary to perform an additional enzymatic or immunological detection step in order to see the non-fluorescent hapten. The signals are then examined using fluorescence microscopy. Fluorochrome, which emits colourful signals at the hybridization site, is a component of the enzymatic detection. The immunological detection approach is based on antibody binding to certain antigens, which is subsequently demonstrated by a coloured histochemical reaction observable under a light microscope or fluorochromes under UV light.
The specificity of the probe-target binding is critical for the effectiveness of FISH assay. The probe is designed to connect solely to specific sequences, which are frequently unique to a specific gene, chromosome, or cell type. Because of this specificity, FISH can be used to identify individual genes or chromosomes, visualize gene expression patterns, and detect chromosomal abnormalities.
Probes are short single-stranded DNA or RNA molecules that have been fluorescently tagged. These probes are programmed to bind to specific DNA or RNA sequences in a sample. Because of the dye’s fluorescence, the location of the target sequence can be observed and studied. It has a property of hybridizing through complementary base pairing, resulting in the formation of double-stranded hybrids. The probes emit a fluorescence signal when they bind to their target, which can then be visualized under a microscope.
There are different types of probes that can be used in FISH. Probes can be designed to target specific sequences based on the genetic information available for the organism being studied. The probes can be made from either synthetic or naturally occurring DNA or RNA. In order to maximize specificity, probes are often designed to be complementary to the target sequence, but not to other regions of the genome.
Locus specific probes
These probes bind to a specific area of a chromosome. These probes can detect microdeletion and microduplication syndromes within a specific chromosome.
Whole-chromosome probes
These probes are collections of tiny probes tagged with distinct fluorescent dyes, each of which binds to a different sequence throughout a given chromosome. These probes can be used to map the entire chromosome, providing information on chromosomal abnormalities. Scientists can label each chromosome in its own distinct colour by using numerous probes labeled with a mixture of various fluorescent dyes which is known as a spectral karyotype that includes full-colour map of the chromosome.
Repetitive probes
The repeating sequences located in the centre of each chromosome are used to create repetitive probes, which can be used to examine whether a person has the appropriate number of chromosomes. These probes can be used in conjunction with locus-specific probes to identify whether a person lacks genetic material from a given chromosome.
In FISH, fluorescent probes are used to visualize specific DNA or RNA sequences in a sample. There are several types of fluorescent probes that can be used, including:
- Cy3: A green fluorescent dye that is commonly used in FISH experiments.
- Cy5: A red fluorescent dye that is also commonly used in FISH experiments.
- FITC (fluorescein isothiocyanate): A green fluorescent dye that is often used in FISH experiments.
- Texas Red: A red fluorescent dye that is commonly used in FISH experiments.
- Rhodamine: A red fluorescent dye that is sometimes used in FISH experiments.
- DAPI (4′,6-diamidino-2-phenylindole): A blue fluorescent dye that is often used to stain DNA in FISH experiments.
- Alexa Fluor dyes: A family of fluorescent dyes that are available in a wide range of colors, including green, red, and blue. Alexa Fluor dyes are commonly used in FISH experiments.
The steps in Fluorescence In Situ Hybridization (FISH) technique typically include:
Sample preparation
Obtain a biological sample, such as tissue or cells, and prepare it for FISH by fixing and permeabilizing the cells. The sample (e.g., tissue section or cells) is fixed to a microscope slide and treated to preserve the target DNA/RNA and to make the chromosomes visible under a microscope. The slides can be kept in a freezer at 80 °C for at least a year.
Preparation of probes
The probes are single-stranded DNA or RNA sequences complementary to the target sequences. The probes are labeled with a fluorescent dye to make them visible under a fluorescence microscope.
Probe labeling
Label the FISH probes with a fluorescent dye, which will bind to specific regions of the target DNA. Probes can either be directly fluorophore-labeled or indirectly labeled with a hapten that can later be detected via an enzymatic or immunological detection technique.
There are now several techniques for tagging DNA probes for nonradioactive in situ hybridization. The most popular strategy is to label the probe with reporter chemicals (haptens). There are several different haptens on the market, including biotin, digoxigenin, dinitrophenol, fluorescein, rhodamine, AMCA, and coumarin. According to standard protocols, these haptens can be included as labeled nucleotides using the nick translation, random primer labeling, or PCR tagging techniques.
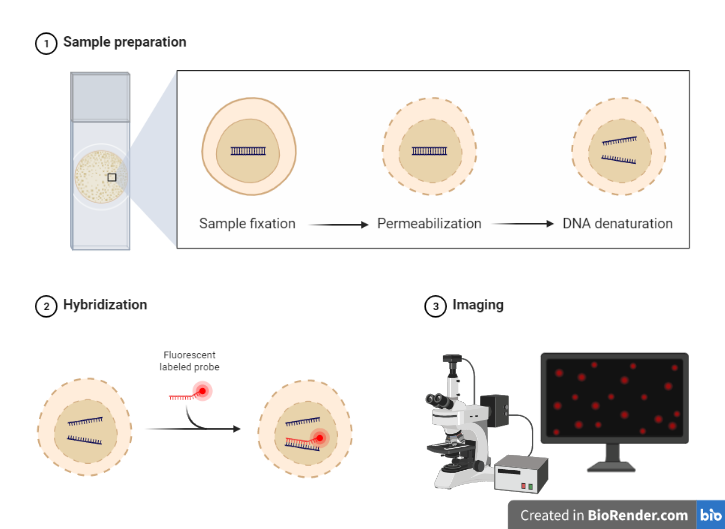
Fig: Steps in FISH Technique
Denaturation: The process of breaking the hydrogen bonds in a double-stranded DNA molecule to create a single-stranded DNA molecule is known as denaturation. Typically, this is accomplished by heating the DNA sample to a high temperature (between 80 and 100 °C), which separates the DNA strands. The complementary probe can then be hybridized with the single-stranded DNA molecules to enable the imaging and localisation of particular DNA sequences within cells or tissues.
Hybridization: Incubate the sample with the labeled probes, allowing them to hybridize (bind) to the target DNA. During hybridization, the labeled probes are added to the sample and are allowed to bind to the complementary DNA sequences in the target. This binding process is specific, meaning that the probes will only bind to their intended target, allowing the researcher to visualize specific regions of the DNA. Hybridization conditions, such as temperature, salt concentration, and incubation time, are carefully controlled to ensure optimal binding of the probes to the target DNA
Washing: Wash the sample to remove unbound probes, reduce background signal, and increase the specificity. A high salt concentration in the washing solution often aids in the removal of unbound probes and the reduction of non-specific binding. Multiple washings of the sample are possible to further drastically reduce background signal.
Detection: Visualize the bound probes using fluorescence microscopy and capture images. Visualization in this technique typically involves the use of fluorescence microscopy to detect and visualize the bound probes. The sample is viewed under a fluorescence microscope equipped with a filter that is specific for the wavelength of light emitted by the fluorescent dye used to label the probes. In the FISH technique, each probe is labeled with a different fluorescence dye, allowing multiple probes to be visualized simultaneously. The fluorescence signals can be viewed in real-time, or captured as images for later analysis.
In addition, the nonfluorescent hapten must first be seen using an enzymatic or immunological detection technique if the probe is indirectly tagged with it.
Analysis: Analyse the captured images to determine the distribution and number of fluorescent signals, which can provide information about genetic changes in the sample.
Interpretation: The results are interpreted and analyzed, taking into account the pattern of fluorescence signals to determine the presence, absence or rearrangement of the target sequences. Interpret the results, which can provide insight into genetic changes, chromosomal aberrations, gene expression patterns, and other biological processes.
Reporting: Report the results, which can be used to guide further research and inform clinical decision making.
Applications:
Prenatal diagnosis of chromosomal abnormalities: It is a widely used technique in cytogenetics to visualize specific DNA sequences in chromosomes and to detect chromosomal abnormalities. FISH can detect specific chromosomal abnormalities, such as deletions, duplications, and translocations, which are often associated with genetic disorders and cancer. In addition, this technique is used in prenatal diagnosis to detect chromosomal abnormalities in a developing foetus, providing important information for genetic counselling and treatment planning.
Detection of copy number variants (CNVs): Copy number variants (CNVs) refer to changes in the number of copies of a specific DNA sequence in a genome. These changes can result in gains or losses of genetic material, which can contribute to genetic disorders and diseases, such as cancer. Fluorescence In Situ Hybridization (FISH) is a widely used technique for the detection of CNVs.
Detection of cancer: FISH is used to diagnose various types of cancer, including leukemia, lymphoma, and solid tumors. The technique can detect specific chromosomal abnormalities and gene rearrangements that are characteristic of certain cancers. It can be used to monitor the response of a cancer to treatment by detecting changes in gene expression and chromosomal structure. This information can help to determine the effectiveness of the treatment and whether a change in treatment is necessary.
Detection of infectious diseases: FISH is used to identify infectious pathogens by analyzing 16S ribosomal RNA (rRNA) that is distinctive to phylogenetic groups. For the investigation of microbial communities in the mouth cavity and gastrointestinal flora, FISH probes made of oligonucleotide sequences (17–34 nucleotides in length) complementary to 16S rRNA can be utilized. The microbial population is highly populated in the intestine and mouth cavity, and FISH has been used to identify pathogens there. The bacteria causing respiratory tract infections have also been targeted in the design of specific oligonucleotide probes. FISH has also been used to identify infections in tissues in a manner similar to that. Pathogenic bacteria in blood cultures have been identified using genus- and species-specific oligonucleotide probes. For instance, FISH probes that are complementary to a certain 16s rRNA sequence can identify malaria infection in blood samples.
Genetics: FISH is used to identify and locate specific genetic mutations in a sample, providing important information for genetic counselling and diagnosis of genetic disorders.
Drug discovery: FISH is used in drug discovery research to evaluate the effectiveness of new drugs by tracking changes in gene expression and chromosomal structure in response to drug treatment.
Neuroscience: FISH is used in neuroscience research to study the localization and expression of specific genes in the brain, providing important insights into the biology of the nervous system.
Limitations:
When using several probes in a single experiment, FISH can be complicated to perform and analyze, requiring specific tools and knowledge. Similar to FISH probes, non-specific targets may bind to them, producing false-positive results and making interpretation more difficult. With an usual resolution of around 10–50 kb, this approach has a relatively low resolution. This implies that it might not be able to recognize tiny CNVs or alterations in the genome’s structural elements.
While FISH is an effective method for identifying particular DNA sequences and chromosomal aberrations, there are some drawbacks that must be taken into account when determining if FISH is the most appropriate method for a given study.
References:
- Henegariu, O., Bray-Ward, P., Artan, S., Vance, G. H., Qumsyieh, M. and Ward, D. C. (2001) ‘Small marker chromosome identification in metaphase and interphase using centromeric multiplex FISH (CM-FISH)’, Laboratory investigation, 81(4), 475-481.
- Parra, I., & Windle, B. High resolution visual mapping of stretched DNA by fluorescent hybridization. Nature Genetics 5, 17–21 (1993)
- Fluorescence In situ Hybridization: Cell-Based Genetic Diagnostic and Research Applications.
- Shakoori AR. Fluorescence In Situ Hybridization (FISH) and Its Applications. Chromosome Structure and Aberrations. 2017 Feb 10:343–67.
Leave a Comment Cancel Reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
This page has been archived and is no longer updated
Fluorescence In Situ Hybridization (FISH)
Cytogenetics entered the molecular era with the introduction of in situ hybridization , a procedure that allows researchers to locate the positions of specific DNA sequences on chromosomes. Since the first in situ hybridization experiments in 1969 (Gall & Pardue, 1969), many variations of the procedure have been developed, and its sensitivity has increased enormously. Today, most in situ hybridization procedures use fluorescent probes to detect DNA sequences, and the process is commonly referred to as FISH (fluorescence in situ hybridization). A variety of FISH procedures are available to cytogeneticists, who use them to diagnose many types of chromosomal abnormalities in patients. The success of FISH, and all other methods of in situ hybridization, depends on the remarkable stability of the DNA double helix .

In Situ Hybridization Is Used to Localize DNA Sequences on Chromosomes
In 1953, James Watson and Francis Crick described the extensive network of hydrogen bonds that hold together the two antiparallel strands in the DNA double helix (Watson & Crick, 1953). Today, even schoolchildren know that adenine on one DNA strand binds to thymine on the complementary DNA strand, and that cytosine likewise binds to guanine . Because of the many hydrogen bonds formed between these bases, the double helix is a remarkably stable structure. Moreover, if the hydrogen bonds that hold the helix together are broken with heat or chemicals, the helix is able to re-form when conditions become more favorable. This ability of the DNA helix to re-form, or renature, provides the basis for molecular hybridization.
In molecular hybridization, a labeled DNA or RNA sequence is used as a probe to identify or quantify the naturally occurring counterpart of the sequence in a biological sample . In the 1960s, researchers Joseph Gall and Mary Lou Pardue realized that molecular hybridization could be used to identify the position of DNA sequences in situ (i.e., in their natural positions within a chromosome ). In fact, in 1969, the two scientists published a landmark paper demonstrating that radioactive copies of a ribosomal DNA sequence could be used to detect complementary DNA sequences in the nucleus of a frog egg . Since those original observations, many refinements have increased the versatility and sensitivity of the procedure to the extent that in situ hybridization is now considered an essential tool in cytogenetics.
Fluorescent Probes Are Introduced
Soon after Gall and Pardue's work, fluorescent labels quickly replaced radioactive labels in hybridization probes because of their greater safety, stability, and ease of detection ( Rudkin & Stollar, 1977 ). In fact, most current in situ hybridization is done using FISH procedures ( Trask, 2002 ; Speicher & Carter, 2005 ). Detecting a DNA sequence can be compared to looking for a needle in a haystack, with the needle being the DNA sequence of interest and the haystack being a set of chromosomes. This search is made much easier if the investigator has a powerful "magnet"—in this case, a fluorescent copy of the DNA sequence of interest. Hybridization occurs when the "magnet" meets the "needle"; this requires both a probe and a target, as shown in Figure 1. In the figure, the probe sequence, often a piece of cloned DNA, is shown in red. The target DNA—chromosomes on a glass slide—is shown in blue (in the right column). Hydrogen bonds that join the two strands of the DNA helix are represented by black lines.
The first step in the process is to make either a fluorescent copy of the probe sequence (Figure 1b, middle column) or a modified copy of the probe sequence that can be rendered fluorescent later in the procedure (Figure 1b, left column). Next, before any hybridization can occur, both the target and the probe sequences must be denatured with heat or chemicals (Figure 1c). This denaturation step is necessary in order for new hydrogen bonds to form between the target and the probe during the subsequent hybridization step. The probe and target sequences are then mixed together (Figure 1d), and the probe specifically hybridizes to its complementary sequence on the chromosome. If the probe is already fluorescent (middle column), it will be possible to detect the site of hybridization directly. In other cases (left column), an additional step may be needed to visualize the hybridized probe. Hybrids formed between the probes and their chromosomal targets can be detected using a fluorescent microscope.
When investigators design a FISH experiment, they need to consider whether the sensitivity and resolution needed for the experiment lie within the technical limits of fluorescence microscopy. Sensitivity depends on the light-gathering ability of the particular microscope, which determines whether small target sequences, which are more difficult to see than large target sequences, can be detected. Resolution refers to the ability to distinguish between two points along the length of a chromosome. Ultimately, light microscopy cannot resolve objects that are separated by less than 200–250 nm, the lower limit of the visible light spectrum. With these technical limits in mind, investigators also need to consider the conformation of DNA within the chromosome. Metaphase chromosomes are thousands of times more compacted than interphase chromosomes, which in turn are at least ten times more compacted than naked DNA. (Remember that one 3.4 nm turn of the DNA helix corresponds to 10 base pairs of DNA.) When all these factors are considered together, investigators typically expect to obtain resolution in the range of megabases for positions on metaphase chromosomes and resolution in the range of tens of thousands of kilobases for interphase chromosomes.
Using FISH to Identify the Positions of Genes
FISH provides a powerful tool for identifying the location of a cloned DNA sequence on metaphase chromosomes. Figure 2a shows the results of a typical FISH experiment, in which a cloned DNA sequence was hybridized to normal metaphase chromosomes. Red bands are detected at hybridization sites on two homologous chromosomes, which can be identified by their characteristic banding patterns. Closer examination shows that each red band actually consists of two spots, corresponding to the two sister chromatids in a mitotic chromosome. A skilled cytogeneticist would be able to use these hybridization data together with the banding pattern to place the probe sequence within a few megabases of other known genes on the chromosome.
Diagnosing Chromosomal Abnormalities Using Karyotypes and FISH
FISH and other in situ hybridization procedures are important in the clinical diagnosis of various chromosomal abnormalities, including deletions, duplications, and translocations. Figure 2b shows one example in which investigators used FISH together with standard karyotyping to analyze a patient translocation . The hybridization probe corresponded to a segment of chromosome 19 that was suspected to include the translocation breakpoint. Three areas of hybridization are apparent in the fluorescent image. One spot corresponds to the patient's normal copy of chromosome 19 (nl19), and the other two spots correspond to the altered, or derived (der), versions of chromosomes 11 and 19 that were produced during the translocation. Thus, investigators were able to use the data both to narrow down the breakpoint region on chromosome 19 and to identify the second chromosome involved in the translocation.
The hybridization probe used in Figure 2b was one of thousands of bacterial artificial chromosome (BAC) clones from the HGP that have been made available to the scientific community. Today, cytogeneticists are able to use extensive HGP clone resources to precisely identify the sites of chromosomal rearrangements that appear in karyotypes. In fact, a consortium of scientists has mapped over 7,000 DNA clones from the HGP to specific bands on human chromosomes ( BAC Research Consortium, 2001 ). At least one clone is available for every megabase segment of chromosomal DNA. (The only exception is the Y chromosome , because it is relatively gene-poor.)
Using Collections of FISH Probes to “Paint” Entire Chromosomes
With multifluor FISH, investigators first prepare a collection of DNA sequences to be used as probes for each chromosome. In Figure 3a, the probe chromosomes have been physically separated from one another by flow cytometry. (Today, investigators would probably use commercially available DNA collections for each chromosome.) In the next step, the DNA samples are labeled with combinations of fluorochromes that produce a unique color for each chromosome. (The Cot-1 DNA step in the figure removes repetitive DNA sequences [e.g., centromeric DNA] that would bind to all chromosomes.) The fluorescent hybridization probes are then combined with and hybridized to metaphase chromosomes. Figure 3b shows images of interphase and metaphase chromosomes as they would appear through a microscope after hybridization. To human eyes, several of the metaphase chromosomes appear to have the same color, but digital processing of the image would distinguish spectral differences between the chromosomes. A normal human chromosome (Figure 3b) will have a uniform color along its length, but a rearranged chromosome will have a striped appearance.
Although chromosome paints allow rapid assessment of large chromosomal changes in metaphase spreads, the resolution of the method is limited. Thus, while chromosome painting allows investigators to quickly identify chromosomes involved in translocations and to identify large deletions and/or duplications, small deletions and duplications will not be detectable. If investigators need more detailed information about the actual sequences involved in chromosomal rearrangements, they need to follow up with site-specific probes, as previously described (Figure 2).
Using FISH to Analyze Interphase Chromosomes
Since the introduction of FISH, cytogeneticists have been able to analyze interphase chromosomes as well as the metaphase chromosomes used in karyotypes ( Trask, 2002 ). This offers a real practical advantage, in that cells do not need to be cultured for several days or weeks before chromosomes can be prepared for analysis. In addition, FISH can be used to analyze chromosomes from specimens such as solid tumors, which are of great clinical interest but do not divide frequently. Another useful feature of FISH is that researchers are able to simultaneously monitor multiple sites if the hybridization probes have been labeled with different fluorophores.
Figure 4 shows two examples of how interphase FISH can be used to diagnose chromosome abnormalities. Figure 4a shows an interphase nucleus from a patient with Charcot-Marie-Tooth disease (CMT) type 1A (Lupski et al. , 1991). CMT type 1A is a relatively common neurological condition caused by a duplication in a gene on chromosome 17 that encodes one of the proteins in the myelin sheath that surrounds nerve axons. In Figure 4a, the patient's cell has been hybridized with a red-labeled probe corresponding to a sequence within the duplicated region, along with a green probe corresponding to a sequence on chromosome 17 that lies outside of the duplicated region. From the two green signals, it is possible to locate two copies of chromosome 17 within the nucleus. One chromosome has the normal configuration , while the second, der(17), contains the duplicated region, which is evident from two nearby red signals. The figure also serves to illustrate another important feature of interphase FISH. Because interphase chromatin is about 10,000 times less compacted than mitotic chromatin, it is possible to resolve the duplicated regions on der(17) as discrete points. This small duplication would have been difficult to resolve in mitotic chromosomes.
Figure 4b shows a FISH analysis that was used to detect the presence of a chromosomal translocation in a patient suffering from chronic myelogenous leukemia (Tkachuk et al. , 1990). In most cases of this disease, a segment of chromosome 9 that contains the ABL proto-oncogene fuses with the breakpoint cluster region ( BCR ) on chromosome 22 during a reciprocal translocation . The derived chromosome 22, or der(22), also known as the Philadelphia chromosome, contains a BCR-ABL fusion gene in which the powerful BCR promoter drives synthesis of the ABL oncogene transcript, leading to cancer . Figure 4b demonstrates that BCR-ABL fusions can be readily identified by FISH when a green-labeled hybridization probe flanking BCR is applied together with a red-labeled probe flanking ABL . In this image, the normal copies of chromosomes 9 and 22 are detected as red and green spots, respectively. On the other hand, the Philadelphia chromosome is visible as a complex fused spot, which appears to have a central yellow region with red and green subregions on either side. (In fluorescence microscopy, yellow is indicative of very close proximity of red and green probes, such that they appear to overlap.) The intricate substructure of the fused spot is detectable in interphase chromosomes, but it would not be resolved in a similar FISH analysis of metaphase chromosomes. Thus, two-color interphase FISH provides a sensitive method for analyzing chromosome fusion events without the need for a prior cell culture .
Another research application of interphase FISH makes use of chromosome-specific paints to obtain information about the organization of chromosomes within the nucleus. Figure 2a (upper left) shows an interphase nucleus that has been stained with chromosome-specific paints. One can see from the figure that the chromosomes occupy distinct territories within the nucleus. By creatively combining chromosome-specific probes with gene-specific probes and antibodies, investigators can use FISH to provide exciting new insights about nuclear architecture.
Additional Applications of FISH in the Clinic and Research Laboratory
Exciting new applications of FISH that extend its range continue to be developed. For example, cytogeneticists now use comparative genomic hybridization to detect quantitative differences, like copy number variations, in the chromosomes of their patients. Recently, investigators have also been able to increase the resolution of FISH by using stretched chromatin fibers (Parra & Windle, 1993) or microarrays as the target. With tools such as these, cytogenetics has been able to move from studying whole chromosomes on the macroscopic scale , to studying the DNA of which these chromosomes consist.
References and Recommended Reading
- Add Content to Group
Article History
Flag inappropriate.


Email your Friend

- | Lead Editor: Clare O'Connor

Within this Subject (43)
- Chromosome Analysis (10)
- Chromosome Number (4)
- Chromosome Structure (6)
- Chromosome Theory and Cell Division (5)
- Mutations and Alterations in Chromosomes (13)
- Sex Chromosomes (5)
Other Topic Rooms
- Gene Inheritance and Transmission
- Gene Expression and Regulation
- Nucleic Acid Structure and Function
- Chromosomes and Cytogenetics
- Evolutionary Genetics
- Population and Quantitative Genetics
- Genes and Disease
- Genetics and Society
- Cell Origins and Metabolism
- Proteins and Gene Expression
- Subcellular Compartments
- Cell Communication
- Cell Cycle and Cell Division
© 2014 Nature Education
- Press Room |
- Terms of Use |
- Privacy Notice |

Visual Browse
- Helpful Links
That Fish Blog – Aquarium Advice and Information
Aquatic science fair projects for aquarium lovers.
Posted by: Eileen Daub in Common Aquarium Questions , Do-It-Yourself , Fish Health , Fun Stuff September 13, 2014 0 25012 Views
With the new school year getting underway, it’ll only be a matter of time before the first science fairs and lab experiments start up. We get lots of students visiting or contacting us in search of ideas and test subjects for project ranging from the simple to the complex so I thought I’d share some tips and some of our favorite ideas for easy (and affordable) aquatic science fair projects for students of all levels.
Before you begin…
Before you start any experiment or project involving live plants or animals, it is important to make sure you are prepared for the maintenance, care and cost of the experiment and livestock they are taking on. We’ve had many students that have a great idea for a project involving crabs, jellyfish, “Nemo” clownfish or other animals but don’t realize how much work and supplies are going to be needed to keep these animals healthy. They are also a commitment after the experiment is done; fish aren’t disposable and you have to be ready for them to go from Test Subject to Pet at the end of the project. Some experiments also require multiple subject that each need their own setup and supplies.
Time is another important factor to consider with these projects. Many experiments involving fish and plants are going to take some time before you will really start to see results like growth or changes to color or behavior. It can take at least a few weeks to reach any conclusions so if this is a last-minute project, you might be better off heading in another direction.
The hypothesis of an experiment is the question you are trying to answer or what you are trying to prove or study. Depending on your grade level, this might be observing an environment or behavior or it might be testing a number of variables against a control. If one of the ideas here intrigues you but you aren’t sure where to go with it, ask your teacher or let us know and we might be able to help point you in a direction.
The project ideas here are mainly freshwater. Saltwater science fair projects are definitely possible, but they are going to be more difficult and expensive. Saltwater aquariums have a lot more factors than freshwater and need more equipment and maintenance to keep them healthy. If you already have a saltwater aquarium, you may be able to adapt one of these ideas to it or come up with your own but it may be easier to stick with a freshwater experiment if you don’t already have some saltwater experience.
Some Aquatic Science Project Ideas
A male Veiltail Betta
The Betta (“Siamese Fighting Fish”) as a test subject
- Betta splendens , also known as the Siamese Fighting Fish, is a good fish to use in aquatic studies and projects. They don’t need a lot of equipment or space, are easy to care for and are fairly inexpensive. They can be found in a lot of different colors and varieties but the most common kind that you will find in most stores is the Veiltail Betta . Some project ideas would be keeping bettas in a few different environments like a brightly lit versus darkened room, under a colored light like blue (“actinic”) light versus a white light, or warmer versus cooler temperatures. How do those changes affect each fish’s growth or coloration? You can take a picture every few days to compare any changes in their color or fins. Does the type of food or the color of the ornaments and backgrounds compared to the fish’s own color make any difference? Try different types or brands of food (flakes, pellets, frozen food, live food, freeze-dried food) to see how each affects the fish’s health.
- A good way to measure growth is to weigh a specific amount of water in a container without the fish, then weigh the same container with the fish. The difference between the two weights will tell you about how much the fish itself weighs and changes in that weight will tell you if the fish has gained or lost weight.
Fish Food Nutritional Comparison
Color-enhancing flake food like this is a common aquarium food.
- Anyone who has gone shopping for food for their aquarium knows how many choices there are for your fish. Flakes, pellets, frozen, freeze-dried, live, color-enhancing, vitamin-enhanced…the choices can be overwhelming. At the time that I write this, we have over 400 different fish food items on our website alone! For a science fair project, you can compare several of these foods and see how your fish react; feed the same kind of fish in separate but identical environments the same amount of different types or brands of food at the same time of day and measure how this impacts their growth, health and behavior. Maybe one kind of food is advertised as color-enhancing or another has a different primary ingredient…does one kind give the fish brighter colors or make them grow more during the time frame of the experiment? The fish should all be in separate aquariums to make sure they are only eating “their” food but you can otherwise use any fish – goldfish, bettas, livebearers like mollies or guppies. You can use the same method as above to record their growth.
The Nitrogen Cycle
The Nitrogen Cycle and “Cycling” a New Aquarium
- The Nitrogen Cycle is a process that every aquarium and contained body of water will go through as the helpful bacteria populations that take care of fish waste will go through. Every aquarist has seen this happen whether they realize it or not. As the first living critters in a tank produce waste or leftover food or other material decomposes in the tank, a bacteria known as Nitrosomonas change that waste from ammonia (NH 3 ) to nitrite (NO 2 – ). Another population known as Nitrobacter will change that Nitrite into Nitrate (NO 3 – ), the end product of the cycle which is usually removed by water changes or plants that use it to grow like fertilizer. If the levels of Ammonia, Nitrite and Nitrate could be put on a graph throughout the process, they should look like three hills with their peaks coming one after another. Can you set up and “cycle” a new aquarium and use test kits to record these values at a specific interval (every 5 hours, for example) and see if you can recreate this cycle? Does the number of fish change the levels or speed throughout this cycle? What about if the tank is cycled with a dead “waste” like leftover food instead of live fish?
- A project like this does use chemical test kits. The kits available from pet stores for aquariums are easy to use, but adult supervision should always be used around chemicals like these.
Fish Tricks
- Did you know fish can learn? Fish like goldfish have been “trained” to recognize objects and do all sorts of tricks like swimming through hoops or under limbo bars. There have even been “soccer matches” between saltwater fish to celebrate the World Cup. Training fish, like most animals, is all about positive reinforcement. Some companies have even made training kits just for teaching fish . Some experiments have trained fish to recognize certain colors by putting food in cups of one color (like red) but not others (like blue and yellow). Once the fish begins to associate the red cup with food, does putting a red ornament in the tank or red background on part of the tank make them choose that over the blue or yellow? How long does it take, and does any kind of fish learn faster than others? You can try putting one fish through its paces or train a few fish to compare their learning styles.
- Check out this video from Rochester Institute of Technology’s Professor Caroline DeLong to see how she taught a goldfish to recognize shapes
Brine Shrimp Hatchery Kit
Hatching Brine Shrimp
- Brine Shrimp are popular as fish food in aquariums as well as in the wild, and there was a time that “Sea Monkeys” were popular as “pets”. Hatching brine shrimp can be an interesting endeavor. They can be easy to grow but need just the right salinity (salt level) and the newly-hatched shrimp will need equally tiny food. The baby brine shrimp also show a behavior known as “phototaxis”, meaning they are attracted to light. Brine shrimp eggs are inexpensive and can be purchased in their dry “cyst” form where they will hatch once back in the right water. Experiment with salt levels, food like phytoplankton and with light and see what factors make for the best hatches. Once the shrimp are hatched, they can grow to almost a half inch in length and make great fish food!
This is just a sampling of some concepts that aquarists face every day and there are a lot more out there like overfeeding , breeding and genetics in fish like guppies or mollies, water changes and water chemistry…the possibilities are endless! Popular aquarium fish are even ending up in important medical research . Have we given you any ideas for your next science fair project or laboratory experiment? What would you like to learn more about, and what are some of your favorite projects? Share your results and experiences with us…we’d love to see the results of your studies!
About Eileen Daub
Read other posts by Eileen Daub
Related Articles
Dip treatments for fish and corals.
June 9, 2021
Why We Wait Until May 1st to Sell Pond Fish & Plants
April 21, 2017
Keeping Finding Dory Characters in the Home Aquarium
June 15, 2016

Fish Behavior Science Fair Ideas

The natural world is full of wonder and mystery, making for entertaining and enlightening science projects. Experimenting on fish, in particular, can make for a winning science fair project that is also fun to perform. Whenever a budding scientist works with animals, the utmost care should be taken to prevent the creatures from experiencing undue harm.
Goldfish Memory
An common myth is that goldfish have comically short memory spans. Put this myth to the test by purchasing several goldfish and placing them all in the same tank. Use non-toxic, waterproof paint to color one thimble red and another thimble blue. Each day, put the fish's food in the red thimble and slowly lower both thimbles into the fish tank with fishing line. If the goldfish begin to dart toward the red thimble to obtain the food within, you could deal a death blow to this myth. Be sure to insert the thimbles into different parts of the tank each day to ensure that the fish are responding to color instead of thimble placement.
Effect of Water Temperature on Respiration
Perform this experiment to determine if water temperature affects fish respiration. Place several fish in a tank with the water at the prescribed temperature, which can be discerned from knowledgeable pet store employees or online resources. Over the course of one week, count twice daily how many times each fish breathes per minute (look for fluttering gills and closing mouths when the fish takes a breath). Use a stopwatch to guarantee an accurate count and carefully record your results in a journal or notebook. The following week, raise the water temperature five degrees with an aquarium heater. Record respiration rates and compare to your original results. Raise the temperature a few more degrees for even more results, but do not raise the temperature so high as to injure or kill the fish. Ask a knowledgeable pet store employee or search online resources to discover how hot is too hot for your particular species of fish.
Affect of Light on Behavior
Purchase several small fish tanks and insert ten fish of the same species into each. Outfit each tank with a different light, such as regular fluorescent bulbs, compact fluorescent bulbs, LED aquarium bulbs and black light bulbs. For several weeks, record and compare the behavior of the fish in a journal at least twice a day. Observe their reaction to movement near the tank, their proximal relationships to each other, how much they eat, how quickly they eat and their general amount of movement. Make sure that you keep all other variables constant, such as food amount and water temperature.
Affect of Sound on Behavior
Purchase three small fish tanks and insert an equal number of fish into each. Place one tank in a quiet room, and for the next few weeks, ensure that the fish live in a silent environment (no talking or music). Place another tank in a room outfitted with a stereo that constantly plays music of different types. Place the final tank in another room and, as you feed them, talk to the fish. Observe and compare the fish behavior in a journal at least twice a day for several weeks to determine if sound affects fish behavior. Observe their reaction to movement near the tank, their proximal relationships to each other, how much they eat, how quickly they eat, and their general amount of movement.
- Julian Trubin: Fish and Goldfish Science Fair Ideas
- Fish Lore: Aquarium Lighting
Cite This Article
Kopf, Aaron. "Fish Behavior Science Fair Ideas" sciencing.com , https://www.sciencing.com/fish-behavior-science-fair-ideas-6391872/. 22 November 2019.
Kopf, Aaron. (2019, November 22). Fish Behavior Science Fair Ideas. sciencing.com . Retrieved from https://www.sciencing.com/fish-behavior-science-fair-ideas-6391872/
Kopf, Aaron. Fish Behavior Science Fair Ideas last modified August 30, 2022. https://www.sciencing.com/fish-behavior-science-fair-ideas-6391872/
Recommended

- Phage Display Library Construction
- Phage Display Library Screening
- Yeast Display Library Construction
- Yeast Display Library Screening
- Premade Antibody Library
- Premade Peptide Library
- Premade SdAb Library
- Immune Human Antibody Library
- Premade Antibody Library Screening
- Premade Single Domain Antibody (SdAb) Library Screening
- Premade Peptide Library Screening
- Single B Cell Sorting based Antibody Discovery
- Hybridoma based Antibody Discovery
- Transgenic Mice based Human Antibody Discovery
- Yeast Two-Hybrid (Y2H)
- Bacterial Two-Hybrid (B2H)
- SdAb Library Construction & Screening
- Human/Humanized SdAb Discovery
- Bispecific SdAb Discovery
- Monoclonal Antibody Discovery
- Polyclonal Antibody Discovery
- Scaffold Discovery
- Peptide Discovery
- Membrane Protein Antigen Preparation
- GPCR Specific Antibody Discovery
- Ion Channel Specific Antibody Discovery
- Transporter Specific Antibody Discovery
- Membrane Protein Specific Single Domain Antibody (SdAb) Discovery
- Phosphorylation Specific Antibody Discovery
- Acetylation Specific Antibody Discovery
- Methylation Specific Antibody Discovery
- Ubiquitination Specific Antibody Discovery
- Fatty Acylation (Myristoylation, Palmitoylation) Specific Antibody Discovery
- FREE Antibody recognized Anti-ID Antibody Discovery
- TOTAL Antibody recognized Anti-ID Antibody Discovery
- BOUND Antibody recognized Anti-ID Antibody Discovery
- T Cell Receptor recognized Anti-ID Antibody Discovery
- Idiotype Specific Single Domain Antibody (SdAb) Discovery
- Hapten Specific Antibody Discovery
- Thermal Stable mAb Discovery
- Thermal Stable ScFv/Fab Discovery
- Thermal Stable SdAb Discovery
- pH-Sensitive Binder Discovery
- Internalizing Antibody Discovery
- SdAb based Intrabody Discovery
- Agonistic/Antagonistic Antibody Discovery
Creative Biolabs is offering the most comprehensive services for antibody development projects. With strict regulation and effective execution, we are dedicated to providing the most valuable solutions to complete your projects.
- Antibody Affinity Measurement
- Membrane Protein Affinity Measurement
- Small Molecule Affinity Measurement
- Paratope Mapping
- Epitope Binning
- B Cell Epitope Mapping
- T Cell Epitope Discovery
- Antibody Off-Target Identification & Specificity Profiling
- Human/Humanized Antibody Sequencing
- Biosimilar mAb Sequencing
- Single Domain Antibody (SdAb) Sequencing
- Rodent Antibody Sequencing
- Rabbit Antibody Sequencing
- Chicken IgY Sequencing
- T Cell Receptor (TCR) Sequencing
- Hybridoma Sequencing
- In Silico Assessment"> In Silico Assessment
- Ex Vivo Assessment"> Ex Vivo Assessment
- In Vivo Assessment"> In Vivo Assessment
- Anti-Drug Antibodies (ADA) Assay
- BCR Repertoire Analysis
- TCR Repertoire Analysis
- HLA Tissue Typing
- B Cell ELISpot
- T Cell ELISpot
- De-immunization
- ITC Assessment
- DSC Assessment
- Functional Analysis
- Humanization (Convert to Human Ab)
- Camelization (Convert to Single Domain Ab)
- Caninization (Convert to Dog Ab)
- Simianization (Convert to NHP Ab)
- Murinization (Convert to Mouse Ab)
- Chimeric Antibody Conversion
- ScFv/Fab Conversion
- Single Domain Antibody (SdAb) Affinity Maturation
- Single Domain Antibody (SdAb) Stability Improvement
- Specificity Optimization
- Cysteine Modification
- T Cell Receptor Engineering
- Fc Engineering
- Labeling/Conjugation
- GPCR Preparation
- Ion Channel Preparation
- Transporter Preparation
- Custom Membrane Protein Production
- Custom Membrane Protein Solubilization and Stabilization
- Custom Membrane Protein QC
- Insect Cell based VLP Production
- Mammalian Cell based VLP Production
- E. coli based VLP Production"> E. coli based VLP Production
- Yeast Cell based VLP Production
- Plant Cell based VLP Production
- Recombinant Antibody Production
- Ascites Fluid based Antibody Production
- Stable Cell Line Construction
- Virtual Prediction
- Amino Acid & Terminal Sequence Analysis
- Phage Display Library Ready-To-Use Kits
- Yeast Display Library Ready-To-Use Kits
- Recombinant Antibody Products
- Full-Length Membrane Protein (Super Nanodisc)
- GPCR Protein
- Transporter Protein
- Ion Channel Protein
- CAR-T Therapy Target Protein
- Biomarkers and CD Protein
- Fc Receptor Protein
- HLA Protein
- Cell-Free Protein
- Other Membrane Protein
- Cell-Free Membrane Protein Expression Kits
- Phospholipid
- Membrane Protein Stable Cell Line Products
- Antibody-Drug Conjugate (ADC) Products
With over a decade of experience in phage display technology, Creative Biolabs can provide a series of antibody or peptide libraries that are available for licensing or direct screening. These ready-to-use libraries are invaluable resources for isolating target-specific binders for various research, diagnostic or therapeutic applications.
- Phage Display Platform
- Yeast Display Platform
- Hybridoma Platform
- Single B Cell Sorting Platform
- Membrane Protein Platform
Creative Biolabs has established a broad range of platforms for developing novel antibodies or equivalents. These cutting-edge technologies enable our scientists to meet your demands from different aspects and tailor the most appropriate solution that contributes to the success of your projects.
- Peer-Reviewed Publications
- Antibody & Antigen FAQ
- Surface Technology FAQ
- Antibody Service FAQ
- Conference Resources
- Support Documents
- Protocol Instances
- Technology Introductions
- Background Knowledges
- Antibody Overviews
- Membrane Protein Overviews
- Scaffold Overviews
- Literature Review
- Surface Display Technology
- Recombinant Antibody
- Therapeutic Antibody
- De Novo Antibody Sequencing"> De Novo Antibody Sequencing
- Membrane Protein
With deep understanding in antibody-related realms and extensive project experience, Creative Biolabs offers a variety of references to help you learn more about our capacities and achievements, including infographic, flyer, case study, peer-reviewed publications, and all kinds of knowledge that can assist your projects. You are also welcome to contact us directly for more specific solutions.

- Licensing and Collaboration
- Distributors
- Reviews & Endorsements
- Scholarship Program
- Corporate Social Responsibility
Get a real taste of Creative Biolabs, one of the most professional custom service providers in the world. We are committed to providing highly customized comprehensive solutions with the best quality to advance your projects.

- Knowledge Hub
- ISH Introduction
- FISH Introduction
"Creative Biolabs is committed to providing highly customized comprehensive solutions with the best quality to advance our global clients’ projects."
Fluorescent In Situ Hybridization (FISH) Introduction

**********************************************************************************************************************************************
What is FISH? [ TOP ]
Fluorescence in situ hybridization (FISH) is a kind of cytogenetic technique which uses fluorescent probes binding parts of the chromosome to show a high degree of sequence complementarity. Fluorescence microscopy can be used to find out where the fluorescent probe bound to the chromosome. This technique provides a novel way for researchers to visualize and map the genetic material in an individual cell, including specific genes or portions of genes. It is an important tool for understanding a variety of chromosomal abnormalities and other genetic mutations. Different from most other techniques used for chromosomes study, FISH has no need to be performed on cells that are actively dividing, which makes it a very versatile procedure.
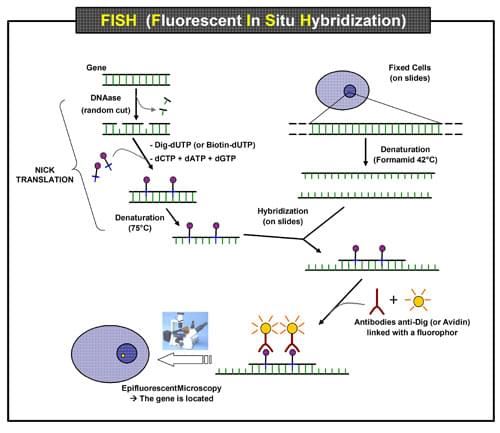
How does FISH work? [ TOP ]
FISH is useful, for example, to help a researcher identify where a particular gene falls within an individual's chromosomes. Here's how it works: o Make a probe complementary to the known sequence. When making the probe, label it with a fluorescent marker, e.g. fluorescein, by incorporating nucleotides that have the marker attached to them. o Put the chromosomes on a microscope slide and denature them. o Denature the probe and add it to the microscope slide, allowing the probe hybridize to its complementary site. o Wash off the excess probe and observe the chromosomes under a fluorescent microscope. The probe will show as one or more fluorescent signals in the microscope, depending on how many sites it can hybridize to.
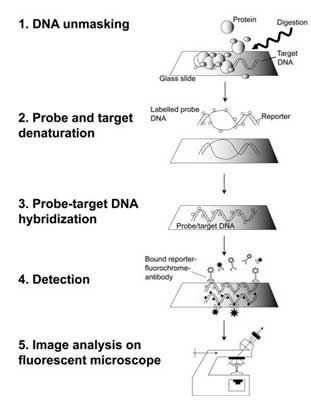
What is FISH used for? [ TOP ]
FISH is widely used for several diagnostic applications: identification of numerical and structural abnormalities, characterization of marker chromosomes, monitoring the effects of therapy, detection of minimal residual disease, tracking the origin of cells after bone marrow transplantation, identification of regions of deletion or amplification, detection of chromosome abnormalities in non-dividing or terminally differentiated cells, determination of lineage involvement of clonal cells, etc. Moreover it has many applications in research: identification of non-random chromosome rearrangements, identification of translocation molecular breakpoint, identification of commonly deleted regions, gene mapping, characterization of somatic cells hybrids, identification of amplified genes, study the mechanism of rearrangements. FISH is also used to compare the genomes of two biological species to deduce evolutionary relationships.
How many types of probes for FISH? [ TOP ]
Generally, researchers use three different types of FISH probes, each of which has a different application: Locus specific probes bind to a particular region of a chromosome. This type of probe is useful when researchers have isolated a small portion of a gene and want to determine on which chromosome the gene is located. Alphoid or centromeric repeat probes are generated from repetitive sequences found in the middle of each chromosome. Researchers use these probes to determine whether an individual has the correct number of chromosomes. These probes can also be used in combination with "locus specific probes" to determine whether an individual is missing genetic material from a particular chromosome. Whole chromosome probes are actually collections of smaller probes, each of which binds to a different sequence along the length of a given chromosome. Using multiple probes labeled with a mixture of different fluorescent dyes, scientists are able to label each chromosome in its own unique color. The resulting full-color map of the chromosome is known as a spectral karyotype. Whole chromosome probes are particularly useful for examining chromosomal abnormalities, for example, when a piece of one chromosome is attached to the end of another chromosome.
Our Services and Products: [ TOP ]
Creative Biolabs offers a full array of custom fluorescence in situ hybridization (FISH) service from probe design, chromosome/cell preparation to expert result interpretation.
Citations/References [ TOP ]
Brandriff, B., Gordon, L. and Trask, B. (1991) 'DNA sequence mapping by fluorescence in situ hybridization', Environmental and molecular mutagenesis, 18(4), 259-262.
Fan, Y.-S., Davis, L. M. and Shows, T. B. (1990) 'Mapping small DNA sequences by fluorescence in situ hybridization directly on banded metaphase chromosomes', Proceedings of the National Academy of Sciences, 87(16), 6223-6227.
Henegariu, O., Artan, S., Greally, J. M., Chen, X.-N., Korenberg, J. R., Vance, G. H., Stubbs, L., Bray-Ward, P. and Ward, D. C. (2001) 'Cryptic translocation identification in human and mouse using several telomeric multiplex fish (TM-FISH) strategies', Laboratory investigation, 81(4), 483-491.
Henegariu, O., Bray-Ward, P., Artan, S., Vance, G. H., Qumsyieh, M. and Ward, D. C. (2001) 'Small marker chromosome identification in metaphase and interphase using centromeric multiplex FISH (CM-FISH)', Laboratory investigation, 81(4), 475-481.
Henegariu, O., Dunai, J., Chen, X.-N., Korenberg, J. R., Ward, D. C. and Greally, J. M. (2001) 'A triple color FISH technique for mouse chromosome identification', Mammalian Genome, 12(6), 462-465.
Henegariu, O., Heerema, N. A., Bray-Ward, P. and Ward, D. C. (1999) 'Colour-changing karyotyping: an alternative to M-FISH/SKY', Nature genetics, 23(3), 263-264.
Henegariu, O., Heerema, N. A., Lowe Wright, L., Bray-Ward, P., Ward, D. C. and Vance, G. H. (2001) 'Improvements in cytogenetic slide preparation: controlled chromosome spreading, chemical aging and gradual denaturing', Cytometry, 43(2), 101-109.
Oliveira, A. M. and French, C. A. (2005) 'Applications of fluorescence in situ hybridization in cytopathology', Acta cytologica, 49(6), 587-594.
Tepperberg, J., Pettenati, M., Rao, P., Lese, C., Rita, D., Wyandt, H., Gersen, S., White, B. and Schoonmaker, M. (2001) 'Prenatal diagnosis using interphase fluorescence in situ hybridization (FISH): 2-year multi-center retrospective study and review of the literature', Prenatal diagnosis, 21(4), 293-301.
FISH Probe samples:
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
- In Situ Hybridization (ISH) Introduction
- Hybridoma Services
- Magic™ Anti-Membrane Protein Antibody Discovery
- Anti-idiotype Antibody Discovery
- Single Domain Antibody (sdAb) Discovery
- Native™ Antibody Discovery
- De Novo Antibody Sequencing
- Antibody Humanization
- Antibody Affinity Maturation
- Post-Translational Modification (PTM) Specific Antibody Discovery
- Hyperdoma™ Platform
- B-Cell Sorting Platform
- Frequently Asked Questions (FAQs)
- Knowledge Center
- Contact us USA Tel: Fax: Email: UK Tel: Email: Germany Tel: Email:

Fish Nutrition and Feeding Experiments
Cite this chapter.

- Tom Lovell 2
925 Accesses
4 Citations
Nutrient requirements cannot be determined unless nutrient consumption by the fish is precisely known. Such studies are conducted in a controlled environment, such as aquaria or tanks, to prevent interaction of environmental effects, such as natural food organisms, temperature, and water quality, with the nutrient variable being studied in the experiment. Some studies, however, which involve evaluation of practical feed formulations or feeding regimes, should be conducted under conditions as similar as possible to the conditions where the results will be applied while at the same time allowing for accurate collection and analysis of data. These studies are conducted in experimental ponds, raceways, pens, and similar enclosures of water.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
- Durable hardcover edition
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Unable to display preview. Download preview PDF.
Similar content being viewed by others
Fish nutrition research: past, present and future.
Multiple feedings enhance the growth performance and feed efficiency of juvenile Litopenaeus vannamei when fed a low-fish meal amino acid-supplemented diet
A broader look at ammonia production, excretion, and transport in fish: a review of impacts of feeding and the environment.
Li, M. and R. T. Lovell. 1992. Comparison of satiate feeding and restricted feeding of channel catfish with different percentages of dietary protein. Aquac. 103: 165–175.
Article Google Scholar
Littell, R. C., A. J. Lewis, and P. R. Henry. 1995. Statistical evaluation of bioavailability assays. In Bioavailability of nutrients for animals, eds. C. B. Ammerman, D. H. Baker, and A. J. Lewis. New York: Academic Press.
Google Scholar
Lovell, R. T., E. W. Shell, and R. O. Smitherman. 1978. Progress and prospects in fish farming. In New protein foods, eds. A. M. Altschul and H. L. Wilke, pp. 261–292. New York: Academic Press.
National Research Council. 1993. Nutrient requirements of fish. Washington, D. C. National Academy of Sciences.
Paripatananont, T. and R. T. Lovell. 1995. Responses of channel catfish fed organic and inorganic sources of zinc to Edwardsiella ictaluri challenge. J. Aquatic Animal Health. 7: 147–154.
Robbins, K. R., H. W. Norton, and D. H. Baker. 1979. Estimation of nutrient requirements from growth data. J. Nutr. 109: 1710–1714.
PubMed CAS Google Scholar
Santiago, C. R. 1985. Essential amino acid requirements of Nile tilapia. Ph.D. dissertation, Auburn University, Auburn, AL.
Shell, E. W. 1963. Effects of changed diets on growth of channel catfish. Trans. Amer. Fish. Soc. 92: 432–434.
Shell, E. W. 1983. Fish farming research. Auburn, A.L: Alabama Agricultural Experiment Station.
Download references
Author information
Authors and affiliations.
Auburn University, Auburn, Alabama, USA
You can also search for this author in PubMed Google Scholar
Editor information
Editors and affiliations, rights and permissions.
Reprints and permissions
Copyright information
© 1998 Springer Science+Business Media New York
About this chapter
Lovell, T. (1998). Fish Nutrition and Feeding Experiments. In: Lovell, T. (eds) Nutrition and Feeding of Fish. Springer, Boston, MA. https://doi.org/10.1007/978-1-4615-4909-3_7
Download citation
DOI : https://doi.org/10.1007/978-1-4615-4909-3_7
Publisher Name : Springer, Boston, MA
Print ISBN : 978-1-4613-7226-4
Online ISBN : 978-1-4615-4909-3
eBook Packages : Springer Book Archive
Share this chapter
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research

IMAGES
COMMENTS
Effective probe design is essential for the success of FISH experiments, ensuring specific and reliable detection of target sequences within cells or tissues. Careful consideration of sequence specificity, length, labeling, and in silico analysis contributes to accurate and meaningful FISH results. Hybridization Process:
Participating in science fair projects is a good way to learn the process of scientific inquiry. By doing such projects, children acquire the skills of discipline, observation and documentation that are critical to experimentation. Science projects on fish are interesting and easy to do. When choosing a project idea, however, it is important you focus on finding a topic that is age-appropriate.
In FISH, fluorescent probes are used to visualize specific DNA or RNA sequences in a sample. There are several types of fluorescent probes that can be used, including: Cy3: A green fluorescent dye that is commonly used in FISH experiments. Cy5: A red fluorescent dye that is also commonly used in FISH experiments.
FISH provides a powerful tool for identifying the location of a cloned DNA sequence on metaphase chromosomes. Figure 2a shows the results of a typical FISH experiment, in which a cloned DNA ...
The Betta ("Siamese Fighting Fish") as a test subject. Betta splendens, also known as the Siamese Fighting Fish, is a good fish to use in aquatic studies and projects.They don't need a lot of equipment or space, are easy to care for and are fairly inexpensive. They can be found in a lot of different colors and varieties but the most common kind that you will find in most stores is the ...
Fish and Goldfish science fair projects and experiments: topics, ideas, resources, and sample projects. Fish & Goldfish Science Fair Projects Ideas and Sample Projects by Grade Level. ... In our experiment, we are trying to determine whether or not this new information is correct.
Repeat these two experiments on two more fish. This study should show fish in non-oxygenated water breathe more times per minute than fish in oxygenated water. Rings of Fish. Like counting a tree trunk's rings for age determination, your student can determine a fish's age by counting the rings on its scales. Ask the employee behind your ...
Experimenting on fish, in particular, can make for a winning science fair project that is also fun to perform. The natural world is full of wonder and mystery, making for entertaining and enlightening science projects. Experimenting on fish, in particular, can make for a winning science fair project that is also fun to perform. ... Perform this ...
Reagents for FISH: Our optimized reagents and solutions will help you perform your FISH experiments more smoothly and efficiently. H. Features: The advantages of our services and products are as follows: o Simple, easy, fast and repeatable o Sensitive o Competitive price o Easy to process multiple slides simultaneously o High quality
the amount of weight the fish will gain during the experiment and assume 1.5 g of dry diet is required for 1 g of weight gain. Thus, if the initial weight of the total number of fish is 1 kg and weight is expected to increase by 500%, the amount of diet needed would equal (1.5)(5)(1) = 7.5 kg.